Isabelle Colas
Dr Isabelle Colas is a geneticist in the Cell and Molecular Sciences group.
Current research interests
During meiosis, homologous chromosome arms can exchange via the recombination mechanism essential for gene shuffling. It starts by the formation of Double-stranded breaks (DSBs), lesions in the arms of chromosomes which initiate gene conversion and crossing. The frequency and the distribution of the DSBs throughout the genome seem to correlate with the frequency and distribution of meiotic recombination events, but DSBs formations are not random, and seem to appear at very specific region of the genome. In wheat, rye and barley, genetic recombination frequency is high near the telomeres, but rare towards the centromeres while in Arabidopsis the recombination seems to be in the region of low chromatin condensation.
Breeding programmes have the objectives to develop more productive and more stable varieties. Hybridization and selection are frequently employed in plant breeding and the success of introgression of special traits such as disease resistance relies on genetic recombination between the host and alien chromosomes. However, because chiasmata are more frequent towards the telomeres, it implies that a large area of the chromosomes rarely recombines. Therefore, a better understanding of chromosome pairing, synapsis and recombination would enable in creating tools for manipulation of meiosis to improve cereal breeding.
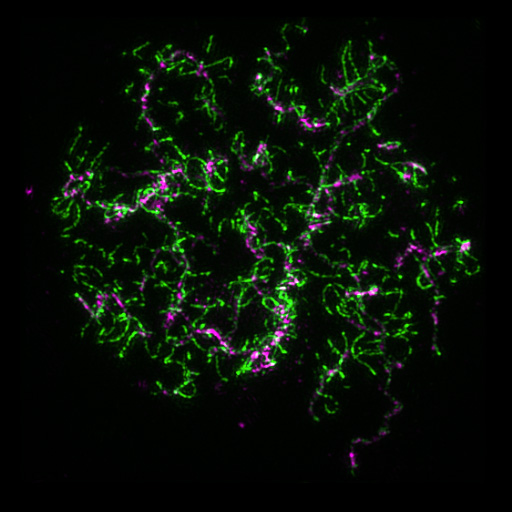 I am especially interested in the cross talk between chromosome synapsis and recombination and their effect on chromatin remodelling. We are currently mapping and characterising a collection of 14 desynapctic mutants first described in early 70s (Hernandes-Soriano, 1973). We are also generating transgenics plant for specific genes of interest to elucidate the mechanism of crossing over control in barley in collaboration with various partner (MeioSys) from the meiosis field and the university of Dundee (A. Barakate, C. Halpin). I am also interested how the chromatin landscape might affect recombination hotspot in barley and within its ancestors (collaboration with A. Flavell).
I am especially interested in the cross talk between chromosome synapsis and recombination and their effect on chromatin remodelling. We are currently mapping and characterising a collection of 14 desynapctic mutants first described in early 70s (Hernandes-Soriano, 1973). We are also generating transgenics plant for specific genes of interest to elucidate the mechanism of crossing over control in barley in collaboration with various partner (MeioSys) from the meiosis field and the university of Dundee (A. Barakate, C. Halpin). I am also interested how the chromatin landscape might affect recombination hotspot in barley and within its ancestors (collaboration with A. Flavell).
Past research
After my university degrees I was determined to work on crop improvement for breeding and make a better food for the future. I started my career in a private company, AGROGENE S.A., specialised in providing molecular markers for breeding program and GMO testing for the worldwide seed market. Supervised by Dr P. Isaac, I was specialised in technical support for our sequencers/analyzer/robot (Applied Biosystems ABI310, 3100, 7700 &7900 and Qiagen robot Q3000) and developing new molecular biology protocols.
I then decided to increase my experience in crop science and started a PhD at the John Innes Centre with Pr G. Moore and P.J. Shaw on the wheat Ph1 locus topic. This is were I developed my main interest in meiosis and the use of cytogenetic techniques to elucidate gene mechanism. I also successfully developed a proteomic approach to study phosphorylation of wheat histone during meiosis and it's affect on chromatin remodelling one of my favourite topic.
Bibliography
- Randall, R. S.; Jourdainb, C.; Nowickac, A.; Kaduchovác, K.; Kubovád, M.; Ayoube, M. A.; Schuberte, V.; Tatoutf, C.; Colasg, I.; Kalyanikrishnab; Dessetf, S.; Mermetf, S.; Boulaflous-Stevensf, A.; Kubalováe, I.; Mandákovád, T.; Heckmanne, S.; Lysakh, M. A.; Panattai, M.; Santoroi, R.; Schubertb, D.; Pecinkac, A.; Routhj, D.; Baroux, C. (2022) Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes, Nucleus, 13(1), 279-301
- Randall, R. S.; Jourdainb, C.; Nowickac, A.; Kaduchovác, K.; Kubovád, M.; Ayoube, M. A.; Schuberte, V.; Tatoutf, C.; Colasg, I.; Kalyanikrishnab; Dessetf, S.; Mermetf, S.; Boulaflous-Stevensf, A.; Kubalováe, I.; Mandákovád, T.; Heckmanne, S.; Lysakh, M. A.; Panattai, M.; Santoroi, R.; Schubertb, D.; Pecinkac, A.; Routhj, D.; Baroux, C. (2022) Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes, Nucleus, 13(1), 279-301
- Wypijewska, D.; Orr, J.; Schreiber, M.; Colas, I.; Ramsay, L.; Zhang, R.; Waugh, R. (2022) The proteome of developing barley anthers during meiotic prophase I, Journal of Experimental Botany, 73(5), 1464-1482
- Darrier, B.; Colas, I.; Gray, G. Rimbert, H.; Choulet, F.; Paux, E.; Bernard, M.; Jenczewski E.; Feuillet, C.; Sourdille, P. (2022) Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number, Plants (Basel), 11(17), Article No. 2281
- Arrieta Salgado, M.; Macaulay, M.; Colas, I.; Schreiber, M.; Shaw, P.; Waugh, R.; Ramsay, L. (2021) An Induced Mutation in HvRECQL4 Increases the Overall Recombination and Restores Fertility in a Barley HvMLH3 Mutant Background, Frontiers in Plant Science, 12, Art. 706560
- Barakate, A.; Orr, J.; Schreiber, M.; Colas, I.; Lewandowska, D.; McCallum, N.; Macaulay, M.; Morris, J.; Arrieta, M.; Hedley, P.E.; Ramsay, L.; Waugh, R. (2021) Barley Anther and Meiocyte Transcriptome Dynamics in Meiotic Prophase I., Frontiers in Plant Science, 11.
- Arrieta, M; Willems, G; DePessemier, G; Colas, I; Burkholz, A; Darracq, A; Vanstraelen, S; Pacolet, P; Barré, C; Kempeneers, P; Waugh, R; Barnes, S; Ramsay, L (2021) The effect of heat stress on sugar beet recombination., Theoretical and Applied Genetics, 134, 81-93
- Orr, J.N.; Waugh, R.; Colas, I. (2021) Ubiquitination in plant meiosis: recent advances and high throughput methods., Frontiers in Plant Science, 12, Article No. 667314
- Pecinka, A.; Chevalier, C.; Colas, I.; Kalantidis, K.; Varotto, S.; Krugman, T.; Michailidis, C.; Vallés, M-P.; Muñoz, A.; Pradillo, M. (2020) Chromatin dynamics during interphase and cell division: similarities and differences between model and crop plants., Journal of Experimental Botany, 71, 5205-5222.
- Arrieta, M.; Colas, I.; Macaulay, M.; Waugh, R.; Ramsay, L. (2020) A modular tray growth system for barley, In: Pradillo M. & Heckmann S. (eds.) Plant Meiosis. Methods in Molecular Biology, Humana, New York, Volume 2061, 367-379
- Darrier, B.; Arrieta, M.; Mittmann, S.U.; Sourdille, P.; Ramsay, L.; Waugh, R.; Colas, I. (2020) Following the formation of synaptonemal complex formation in wheat and barley by high-resolution microscopy, In: Pradillo M. & Heckmann S. (eds.) Plant Meiosis. Methods in Molecular Biology, Humana, New York, Volume 2061, 207-215
- Lewandowska, D.; Zhang, R.; Colas, I.; Uzrek, N.; Waugh, R. (2019) Application of a sensitive and reproducible label-free proteomic approach to explore the proteome of individual meiotic-phase barley anthers., Frontiers in Plant Science, 10, Article No. 393.
- Colas, I.; Barakate, A.; Macaulay, M.; Schreiber, M.; Stephens, J.; Vivera, S.; Halpin, C.; Waugh, R.; Ramsay, L. (2019) Desynaptic5 carries a spontaneous semi-dominant mutation affecting Disrupted Meiotic cDNA 1 (DMC1) in barley., Journal of Experimental Botany, 70, 2683-2698.
- Mittmann, S.; Arrieta, M.; Ramsay, L.; Waugh, R.; Colas, I. (2019) Preparation of barley pollen mother cells for confocal and super resolution microscopy, In: Harwood, W.A. (ed.). Barley: Methods and Protocols. Methods Molecular Biology, Volume 1900, New York: Humana Press, 167-179.
- Colas, I.; Darrier, B.; Arrieta, M.; Mittmann, S.U.; Ramsay, L.; Sourdille, P.; Waugh, R. (2017) Observation of extensive chromosome axis remodeling during the "diffuse-phase" of meiosis in large genome cereals., Frontiers in Plant Science, 8, Article No. 1235.
- Colas, I.; Ramsay, L.; Waugh, R. (2017) Barley meiosis The importance of sticking together., Atlas of Science, 7 March 2017.
- Colas, I.; Baker, K.; Flavell, A.J. (2016) Cytology and microscopy: Immunolocalization of covalently modified histone marks on barley mitotic chromosomes., Bio-Protocol, 6, Article No. e1841.
- Colas, I.; Macaulay, M.; Higgins, J.D.; Phillips, D.; Barakate, A.; Posch, M.; Armstrong, S.J.; Franklin, F.C.H.; Halpin, C.; Waugh, R.; Ramsay, L. (2016) A spontaneous mutation in MutL-Homolog 3 (HvMLH3) affects synapsis and crossover resolution in the barley desynaptic mutant des10., New Phytologist, 212, 693-707.
- Phillips, D.; Jenkins, G.; Macaulay, M.; Nibau, C.; Wnetrzak, J.; Fallding, D.; Colas, I.; Oakey, H.; Waugh, R.; Ramsay, L. (2015) The effect of temperature on the male and female recombination landscape of barley., New Phytologist, 208, 421-429.
- Baker, K.; Dhillon, T.; Colas, I.; Cook, N.; Milne, I.; Milne, L.; Bayer, M.; Flavell, A.J. (2015) Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance., Plant Journal, 84, 111-124.
- Baker, K.; Bayer, M.; Cook, N.; Dreizig, S.; Dhillon, T.; Russell, J.; Hedley, P.E.; Morris, J.; Ramsay, L.; Colas, I.; Waugh, R.; Steffenson, B.; Milne, I.; Stephen, G.; Marshall, D.; Flavell, A.J. (2014) The low recombining pericentromeric region of barley restricts gene diversity and evolution but not gene expression., The Plant Journal, 79, 981-992.
- Barakate, A.; Higgins, J.D.; Vivera, S.; Stephens, J.; Perry, R.M.; Ramsay, L.; Colas, I.; Oakey, H.; Waugh, R.; Franklin, F.C.H.; Armstrong, S.J.; Halpin, C. (2014) The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley., The Plant Cell, 26, 729-740.
- Colas, I.; Tiang, C.; Wang, M.; Sheehan, M.; Pawlowski, W. (2014) Pam1, and RNA Binding Protein required for chromosome dynamics in meiosis., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Ramsay, L.; Colas, I.; Waugh, R. (2014) Modulation of meiotic recombination., In: Kumlehn, J. & Stein, N. (eds.). Biotechnological Approaches to Barley Improvement, Part II. Springer, Berlin, Chapter 16, pp311-329.
- Greer, E.; Martin, A.C.; Pendle, A.; Colas, I.; Jones, A.M.E.; Moore, G.; Shaw, P. (2012) The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat., The Plant Cell, 24, 152-162.
- Randall, R. S.; Jourdainb, C.; Nowickac, A.; Kaduchovác, K.; Kubovád, M.; Ayoube, M. A.; Schuberte, V.; Tatoutf, C.; Colasg, I.; Kalyanikrishnab; Dessetf, S.; Mermetf, S.; Boulaflous-Stevensf, A.; Kubalováe, I.; Mandákovád, T.; Heckmanne, S.; Lysakh, M. A.; Panattai, M.; Santoroi, R.; Schubertb, D.; Pecinkac, A.; Routhj, D.; Baroux, C. (2022) Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes, Nucleus, 13(1), 279-301
- Randall, R. S.; Jourdainb, C.; Nowickac, A.; Kaduchovác, K.; Kubovád, M.; Ayoube, M. A.; Schuberte, V.; Tatoutf, C.; Colasg, I.; Kalyanikrishnab; Dessetf, S.; Mermetf, S.; Boulaflous-Stevensf, A.; Kubalováe, I.; Mandákovád, T.; Heckmanne, S.; Lysakh, M. A.; Panattai, M.; Santoroi, R.; Schubertb, D.; Pecinkac, A.; Routhj, D.; Baroux, C. (2022) Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes, Nucleus, 13(1), 279-301
- Wypijewska, D.; Orr, J.; Schreiber, M.; Colas, I.; Ramsay, L.; Zhang, R.; Waugh, R. (2022) The proteome of developing barley anthers during meiotic prophase I, Journal of Experimental Botany, 73(5), 1464-1482
- Darrier, B.; Colas, I.; Gray, G. Rimbert, H.; Choulet, F.; Paux, E.; Bernard, M.; Jenczewski E.; Feuillet, C.; Sourdille, P. (2022) Location and Identification on Chromosome 3B of Bread Wheat of Genes Affecting Chiasma Number, Plants (Basel), 11(17), Article No. 2281
- Arrieta Salgado, M.; Macaulay, M.; Colas, I.; Schreiber, M.; Shaw, P.; Waugh, R.; Ramsay, L. (2021) An Induced Mutation in HvRECQL4 Increases the Overall Recombination and Restores Fertility in a Barley HvMLH3 Mutant Background, Frontiers in Plant Science, 12, Art. 706560
- Barakate, A.; Orr, J.; Schreiber, M.; Colas, I.; Lewandowska, D.; McCallum, N.; Macaulay, M.; Morris, J.; Arrieta, M.; Hedley, P.E.; Ramsay, L.; Waugh, R. (2021) Barley Anther and Meiocyte Transcriptome Dynamics in Meiotic Prophase I., Frontiers in Plant Science, 11.
- Arrieta, M; Willems, G; DePessemier, G; Colas, I; Burkholz, A; Darracq, A; Vanstraelen, S; Pacolet, P; Barré, C; Kempeneers, P; Waugh, R; Barnes, S; Ramsay, L (2021) The effect of heat stress on sugar beet recombination., Theoretical and Applied Genetics, 134, 81-93
- Orr, J.N.; Waugh, R.; Colas, I. (2021) Ubiquitination in plant meiosis: recent advances and high throughput methods., Frontiers in Plant Science, 12, Article No. 667314
- Pecinka, A.; Chevalier, C.; Colas, I.; Kalantidis, K.; Varotto, S.; Krugman, T.; Michailidis, C.; Vallés, M-P.; Muñoz, A.; Pradillo, M. (2020) Chromatin dynamics during interphase and cell division: similarities and differences between model and crop plants., Journal of Experimental Botany, 71, 5205-5222.
- Lewandowska, D.; Zhang, R.; Colas, I.; Uzrek, N.; Waugh, R. (2019) Application of a sensitive and reproducible label-free proteomic approach to explore the proteome of individual meiotic-phase barley anthers., Frontiers in Plant Science, 10, Article No. 393.
- Colas, I.; Barakate, A.; Macaulay, M.; Schreiber, M.; Stephens, J.; Vivera, S.; Halpin, C.; Waugh, R.; Ramsay, L. (2019) Desynaptic5 carries a spontaneous semi-dominant mutation affecting Disrupted Meiotic cDNA 1 (DMC1) in barley., Journal of Experimental Botany, 70, 2683-2698.
- Colas, I.; Darrier, B.; Arrieta, M.; Mittmann, S.U.; Ramsay, L.; Sourdille, P.; Waugh, R. (2017) Observation of extensive chromosome axis remodeling during the "diffuse-phase" of meiosis in large genome cereals., Frontiers in Plant Science, 8, Article No. 1235.
- Colas, I.; Ramsay, L.; Waugh, R. (2017) Barley meiosis The importance of sticking together., Atlas of Science, 7 March 2017.
- Colas, I.; Baker, K.; Flavell, A.J. (2016) Cytology and microscopy: Immunolocalization of covalently modified histone marks on barley mitotic chromosomes., Bio-Protocol, 6, Article No. e1841.
- Colas, I.; Macaulay, M.; Higgins, J.D.; Phillips, D.; Barakate, A.; Posch, M.; Armstrong, S.J.; Franklin, F.C.H.; Halpin, C.; Waugh, R.; Ramsay, L. (2016) A spontaneous mutation in MutL-Homolog 3 (HvMLH3) affects synapsis and crossover resolution in the barley desynaptic mutant des10., New Phytologist, 212, 693-707.
- Phillips, D.; Jenkins, G.; Macaulay, M.; Nibau, C.; Wnetrzak, J.; Fallding, D.; Colas, I.; Oakey, H.; Waugh, R.; Ramsay, L. (2015) The effect of temperature on the male and female recombination landscape of barley., New Phytologist, 208, 421-429.
- Baker, K.; Dhillon, T.; Colas, I.; Cook, N.; Milne, I.; Milne, L.; Bayer, M.; Flavell, A.J. (2015) Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance., Plant Journal, 84, 111-124.
- Baker, K.; Bayer, M.; Cook, N.; Dreizig, S.; Dhillon, T.; Russell, J.; Hedley, P.E.; Morris, J.; Ramsay, L.; Colas, I.; Waugh, R.; Steffenson, B.; Milne, I.; Stephen, G.; Marshall, D.; Flavell, A.J. (2014) The low recombining pericentromeric region of barley restricts gene diversity and evolution but not gene expression., The Plant Journal, 79, 981-992.
- Barakate, A.; Higgins, J.D.; Vivera, S.; Stephens, J.; Perry, R.M.; Ramsay, L.; Colas, I.; Oakey, H.; Waugh, R.; Franklin, F.C.H.; Armstrong, S.J.; Halpin, C. (2014) The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley., The Plant Cell, 26, 729-740.
- Greer, E.; Martin, A.C.; Pendle, A.; Colas, I.; Jones, A.M.E.; Moore, G.; Shaw, P. (2012) The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat., The Plant Cell, 24, 152-162.
- Colas, I.; Koroleva, O.; Shaw, P.J. (2010) Mass spectrometry in plant proteomic analysis., Plant Biosystems 144, 703-714.
- Colas, I.; Shaw, P.; Prieto, P.; Wanous, M.; Spielmeyer, W.; Mago, R.; Moore, G. (2008) Effective chromosome pairing requires chromatin remodelling at the onset of meiosis., Proceedings of the National Academy of Sciences of the United States of America (PNAS),105, 6075-6080.
- Griffith, S.; Sharp, R.; Foote, T.N.; Bertin, I.; Wanous, M.; Reader, S.; Colas, I.; Moore, G. (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat., Nature, 39, 749-752.
- Arrieta, M.; Colas, I.; Macaulay, M.; Waugh, R.; Ramsay, L. (2020) A modular tray growth system for barley, In: Pradillo M. & Heckmann S. (eds.) Plant Meiosis. Methods in Molecular Biology, Humana, New York, Volume 2061, 367-379
- Darrier, B.; Arrieta, M.; Mittmann, S.U.; Sourdille, P.; Ramsay, L.; Waugh, R.; Colas, I. (2020) Following the formation of synaptonemal complex formation in wheat and barley by high-resolution microscopy, In: Pradillo M. & Heckmann S. (eds.) Plant Meiosis. Methods in Molecular Biology, Humana, New York, Volume 2061, 207-215
- Mittmann, S.; Arrieta, M.; Ramsay, L.; Waugh, R.; Colas, I. (2019) Preparation of barley pollen mother cells for confocal and super resolution microscopy, In: Harwood, W.A. (ed.). Barley: Methods and Protocols. Methods Molecular Biology, Volume 1900, New York: Humana Press, 167-179.
- Ramsay, L.; Colas, I.; Waugh, R. (2014) Modulation of meiotic recombination., In: Kumlehn, J. & Stein, N. (eds.). Biotechnological Approaches to Barley Improvement, Part II. Springer, Berlin, Chapter 16, pp311-329.
- Colas, I.; Tiang, C.; Wang, M.; Sheehan, M.; Pawlowski, W. (2014) Pam1, and RNA Binding Protein required for chromosome dynamics in meiosis., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Colas, I. (2020) Race for barley., James Hutton Institute Barley Away Day, Dunkeld, UK, 6-7 February 2020.
- Colas, I.; Macaulay, M.; Mittmann, S.; Ramsay, L.; Waugh, R. (2019) Using barley mutants to understand meiosis and recombination in cereals., COST INDEPTH SEB Conference, Spain, 8-12 December 2019.
- Ramsay, L.; Colas, I.; Macaulay, M.; Sandhu, A.; Armstrong, S.; Waugh, R. (2019) Desynaptic8 is a spontaneous mutation in X-Ray Repair Cross Complementing 2 (XRCC2) in barley. , British Meiosis Meeting, Aberystwyth, UK, 17-18 April 2019.
- Schreiber, M.; Barakate, A.; Uzrek, N.; Colas, I.; Lewandowska, D.; Macaulay, M.; Mittmann, S.; Arrieta, M.; Clavijo, B.; Wright, J.; Ramsay, L.; Waugh, R. (2018) Developing bioinformatics resources to study meiosis in the barley cultivar Golden Promise., Plant and Animal Genome, Sa Diego, USA, 13-17 January 2018.
- Barakate, A.; Schreiber, M.; Stephens, J.; Davidson, D.; Uzrek, N.; Macaulay, M.; Colas, I.; Lewandowska, D.; Sourdille, A.; Arrieta, M.; Mittmann, S.; Ramsay, L.; Waugh, R. (2018) Targeted mutagenesis of meiotic genes in barley (Hordeum vulgare): Understanding and manipulating meiotic crossovers., Monogram 2018, Norwich, UK, 24-26 April 2018.
- Colas, I.; Mittmann, S.;Macaulay, M.; Ramsay, L.; Waugh, R. (2018) Using barley mutants to understand meiosis and recombination in cereals., British Meiosis Meeting, Brighton, UK, 24-25 April 2018. (Poster)
- Barakate, A.; Stephens, J.; Colas, I.; Schreiber, M.; Lewandowska, D.; Uzrek, N.; Macaulay, M.; Arrieta, M.; Mittmann, S.; Ramsay, L.; Waugh, R. (2017) Understanding and manipulating meiotic crossovers in barley (Hordeum vulgare)., British Meiosis Meeting, Dundee, 19-20 April 2017. Poster
- Lewandowska, D.; Uzrek, N.; Ten Have, S.; Hodge, K.; Colas, I.; Barakate, A.; Ramsay, L.; Waugh, R. (2017) Proteomic profile of barley male meiocytes at early stages of meiosis., British Meiosis Meeting, Dundee, 19-20 April 2017. Poster.
- Mittmann, S.; Ramsay, L.; Macaulay, M.; Waugh, R.; Colas, I. (2017) Discovery of novel E3 ubiquitin ligase by fine mapping and characterization of the barley desynaptic mutant des12., British Meiosis Meeting, Dundee, 19-20 April 2017. Poster.
- Mittmann, S.; Ramsay, L.; Macaulay, M.; Waugh, R.; Colas, I. (2016) Control of meiosis and recombination in barley; Characterization and fine mapping of the barley desynaptic mutant des12., EPSO/FESPB Meeting , Prague, 26-30 June 2016. Poster.
- Baker, K.; Cook, N.; Bayer, M.; Colas, I.; Ramsay, L.; Russell, J.; Flavell, A.J. (2014) Epigenome of barley: Development and optimisation of a ChiP-Seq protocol to investigate histone modification in barley., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Baker, K.; Bayer, M.; Cook, N.; Dreissig, S.; Dhillon, T.; Russell, J.; Hedley, P.; Morris, J.; Colas, I.; Waugh, R.; Steffenson, B.; Milne, I.; Stephen, G.; Marshall, D.; Flavell, A.J. (2014) The peri-centromeric heterochromatin of barley is a permissive environment for gene diversity, expression and evolution., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Macaulay, M.; Colas, I.; Waugh, R.; Ramsay, L. (2014) Mapping and characterization of the barley desynaptic mutants., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Colas, I.; Macaulay, M.; Mittmann, S.; Waugh, R.; Ramsay, L. (2014) Cytological characterization of the barley desynaptic mutants., Plant and Animal Genome XXII, San Diego, California, USA, 12-16 January 2014.
- Colas, I.; Macaulay, M.; Waugh, R.; Ramsay, L. (2013) Fine mapping and characterization of the barley desynpatic des10., EMBO Meiosis Conference, Germany, September 2013.
- Colas, I.; Ramsay, L.; Waugh, R. (2013) A review of the barley desynaptic mutants., Monogram 2013, Dundee University, 18 April 2013.
- Colas, I.; Ramsay, L.; Waugh, R. (2012) Chiamasta formation in barley is determined at early meiosis., UK Plant Science Meeting 2012, Norwich.
- Colas, I.; Ramsay, L.; Waugh, R. (2012) A review of the barley desynaptic mutants., Chromosome Biology, Genome Evolution and Speciation, 11th Gatersleben Research Conference, Germany, 23-25 April 2012.
- Colas, I.; Ramsay, L.; Waugh, R. (2012) A review of the barley desynaptic mutants., 4th British Meiosis Meeting, University of Birmingham, 28 March 2012.
- Colas, I.; Ramsay, L.; Waugh, R. (2011) Control of meiotic recombination in barley., Plant and Animal Genome XIX, San Diego, California, USA, 15-19 January 2011. (Poster)
Scientific Posters / Conferences
| Attachment | Size |
|---|---|
| 894.59 KB | |
| 881.97 KB | |
| 1.07 MB |